Barcode Renamer
(BCR, version 2.3.1.3.)
– a program by Daryl Lafferty, Arizona State University –
NOTE: check out BCRWatcher, our newer, more versatile image renaming program.
When you use a standardize workflow to digitize specimen, it can be very useful to re-name image files, using catalog numbers as their file name. That way the file name uniquely identified a particular specimen (or its label). This process can be facilitated if your collection uses barcodes as catalog numbers
The Barcode Renamer can read a barcode from an image file and use that barcode to rename the file accordingly.
At Arizona State University we are using this program routinely to digitize Vascular Plant Specimens and upload the files in batch to SEINet.
We are still working on establishing a routine workflow also for digitizing lichen specimens.
The download contains a Zip Archive with these two files:
BCR.exe — the executable program; place it into any folder and run it from there; create a shortcut if you use it often.
Readme.txt — some notes on the newest version.
The first time you run the program it will create a file called Config.txt with parameters how you configured the program.
Changes:
- It no longer is necessary select the barcode format; the program will automatically recognize the two most commonly used formats: code 39 & code 128.
- The program generally works equally well now with full-size herbarium sheets and images of labels (= herbarium packets for lichens and/or bryophytes); the “General” option works for both Labels and Full Size Sheets; therefore there is no longer a setting for “Labels”.
- The “Close-up” setting is now only needed when the barcode almost completely fills the entire image.
- Why would you want to take a picture of only the barcode?
The user now can take a close-up of only the barcode and then take several close-ups in sequence of a specimen, showing details like apothecia, sporophytes, etc. When these images are then submitted to the BCR program, the program will first read the barcode and remember this string. Then each subsequent image that does not contain any barcode will be named with the remembered string and a serialized suffix; i.e. if the barcode was ASU12345, then the following images would be ASU12345-a, ASU12345-b, etc. Once the barcode renamer encounters a new image with another barcode, it will remember that that new string and then continue renaming the files with the new barcode and suffices as before. - The program now handles any barcode orientation (although it is still necessary to select the overall specimen orientation “horizontal” vs. “vertical“; once configured it will read the barcode left to right or right to left.
- The program now includes an on-line help that can be accessed via the “Help”-> “Help” menu [this file is accessible online so it can more easily be updated remotely; you will need to be connected to the internet to use the Help]:
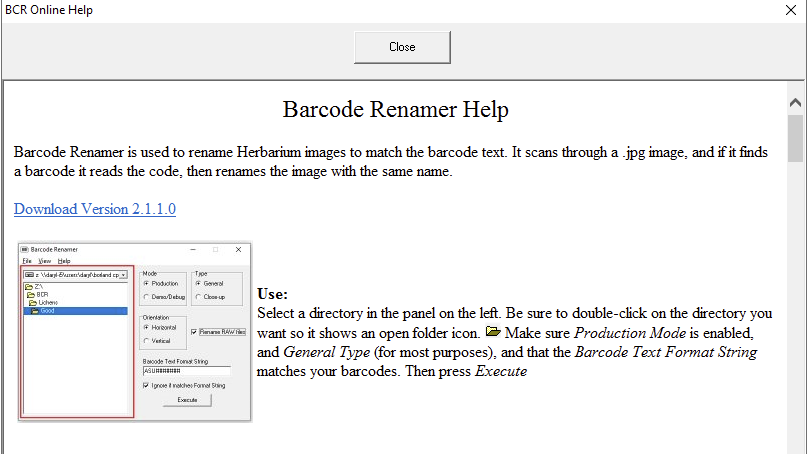
- Overall the program is now more robust, less likely to crash or fail to read.
Using the program to rename image files:
Navigate to the folder, where your image files are located, select the correct barcode position and orientation. If you shoot RAW+JPG pairs, make sure to select the checkbox “Rename Raw Files“.
This program works well with full-size herbarium sheets, but includes options to also rename images of labels and close-up photos. Please send any bug reports and suggestions to CNALH.help@gmail.com
ASU’s Label Digitization Workflow
Here you can also download an example of ASU’s current Herbarium Digitization Workflow. This document describes in detail how we routinely digitize specimen images of Vascular Plant Herbarium Sheets.
This workflow can be easily modified for the digitization of specimen data of lichen or bryophyte packages. It describes the following steps: (1) capturing digital images of labels, (2) using optical character recognition (OCR) to convert the image files into text, and (3) parsing this text into the database fields using Symbiota’s powerful built-in algorithms [please note: the screenshots in this PDF are using SEINet as an example].
