LichenLabler
– a program by Daryl Lafferty, Arizona State University –
Quick Overview: Lichen Labler is a small program that allows you to print labels with barcodes from a CSV directly onto a sheet of paper (letter format) that can then be folded into a a packet (the printout includes folding marks and options to print a barcode).
______________________________________________________________________________________
New in Version 1.12.2 …
Download the Newest Version
Version 1.12.2
The CSV downloaded from a Symbiota portal to print labels currently uses the DarwinCore standard field names, which are not always very intuitive. For example ‘recordedBy‘ is the DarwinCore name for the Collector of a specimen. Or the DarwinCore field ‘recordNumber’ is what is commonly called the ‘Collector’s Number’ (= the number associated with the collector, i.e., the number that uniquely identifies that collection for this particular collector).
Currently, as long as these standard DarwinCore field names are not changed in the download CSV, the program just works ‘out-of-the-box’ with the included configuration file ‘LichenLabelConfig.txt’.
However, if the Symbiota team decides to change these names in the headers of the CSV download, it is now possible to edit the configuration file to map the new field names to their original DarwinCore names.
This is explained in the file ‘LichenLabelConfig.txt’ as follows (any text preceded by // is just a comment, whereas lines without the // are configuration options):
//The program now allows aliases for Darwin Core field names.
//Precede the DarwinCore name with DC_ and follow by a space and then the alias used in your file
//Example: DC_recordedBy Collector
______________________________________________________________________________________
Version 1.10.4
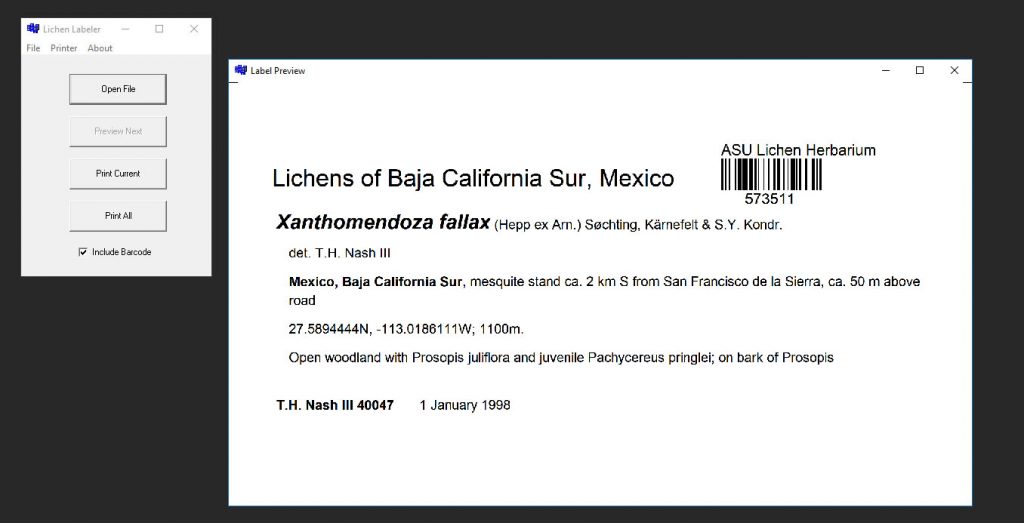
In this version the title “Lichens of …“, “Bryophytes of …” is now centered between the left folding mark and the barcode on the right. This means that even longer titles typically now fit on one line and do not need to wrap into the next. When printing duplicate labels, it is ow possible to modify the text “Duplicate ex …“, for example “Duplicate ex ASU” or “Duplicate ex ASU Lichen Herbarium“, etc. To allow for a longer Duplicate text, this text now appears centered at the bottom of the label. It can be printed in bold or normal font.
Please make sure to use the configuration file ‘LichenLabelConfig.txt’ distributed with this new version, else the program will abort.
Version 1.9.0.0
This version is a significant upgrade. As before you now can print labels on a sheet of paper to be folded into packets (Format: Packets), or you can use it to print regular labels (Format: Labels). You also have the choice whether to print all labels (= Print all) or only the labels for the original specimens (= No Duplicates), or only the duplicates (= Only Duplicates). This is useful, for example, if you want to first print full size packets only for the original specimen (= No Duplicates + Format: Packets) and subsequently regular labels to be placed into the duplicate packets (= Duplicates Only + Format: Labels).
Version 1.9.0.0 also introduces additional formatting configuration options. The LichenLabelConfig.txt file has new optional entries (see configuration below).
Version 1.4.1.0
Bug fix that a label would not print if text was enclosed in double-quotes.
Version 1.4.0.0
The program now works for printing any kind of specimen packet, which means you can change the title of your labels from “Lichens of …” to anything else, e.g., “Bryophytes of …”.
Options to customize labels directly from the CNLAH database are quite limited.
______________________________________________________________________________________
Installation
Extract the files from the Zip Archive into a folder on your System Drive (e.g., C:\LichenLabler). The Zip Archive contains the following files:
- LichenLabeler.exe = the program (double-click to execute)
- LichenLabelConfig.txt = the configuration parameters
- 2018-10-03_ASU_lichen-label_EXAMPLE.csv = an example of data downloaded from the Consortium
- ASU_Example-label.pdf = example standard label packet used at ASU
- ReadMe.txt = a text file with the additions to the configuration options of the new version
______________________________________________________________________________________
Configuration: the newest formatting options (starting version 1.9.0.0.):
The text file LichenLabelConfig.txt can be used to configure the label. The captions preceded by // explain the configuration options in the Text File; below the configuration parameters that we use at ASU.
Please Note: Starting with Version 1.10.4 the last two lines have been added, which allow modification of the duplicate text:
//Parameters below are for Packets
//Distances are from left and top in millimeters
CrossLeft 32 //Left crosses distance from left edge of page
CrossRight 187 //Right crosses distance from left edge of page
CrossTop 90 // Top crosses distance from top of page
CrossBottom 197//Bottom crosses distance from top of page
TitleTop 204 //Distance of the main title from the top of the page (e.g. “Bryphotes of…”)
SciNameTop 214 //Distance of the scientific name from the top of the page
LeftMargin 38 //Distance from the left edge of the page to the text
BarcodeTop 204 //Distance from the top of the page to the top of the barcode
BarcodeLeft 150 //Distance from the left edge of the page to the left edge of the barcode
//
//The next three refer to the four per page labels
FourTop 12 //Distance from top of page to title
FourLeft 8 //Distance from left edge of page to text
FourRight 10 //Distance of right margin from right edge of page
LineSpacing 3.5 //Vertical distance between lines of text
BarcodeHeight 5
FontSize 9 //Standard font
SciNameFont 13 //Font size for scientificName (also bolded and italicized)
TitleFontSize 14 //Font size for main title
Herbarium ASU Lichen Herbarium //Text above the barcode
Prefix Lichens of //Prefix for the main title
TitleBold 1 //0=no, 1=yes
DupText Duplicate ex ASU Lichen Herbarium//change ASU for the name or acronym of your herbarium for your duplicate labels
DupBold 1 //0=no, 1=yes
______________________________________________________________________________________
Configuration: the newest printing options (starting version 1.9.0.0.):
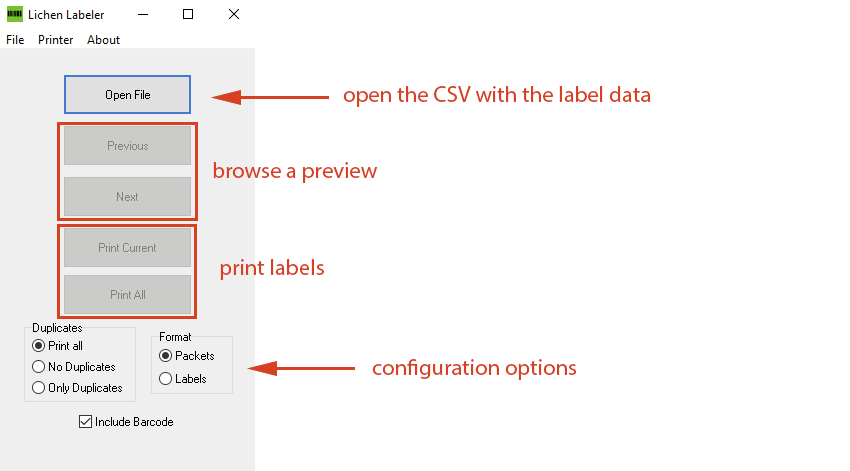
Starting with Version 1.9.0.0 you can now decide whether to print one label per sheet with folding marks (Format: Packets) or four labels per sheet. When printing you can select all entries in your CSV including the duplicates, or only labels for the duplicates, or only the labels of the original specimens (without their duplicates). Even if the ‘Include Barcode’ option is checked duplicates will NOT include barcodes but instead of that barcode display the message ‘Duplicate ex …” with the acronym of the herbarium from which the duplicate specimen originated.
This flexible new configuration options allow you to first print a label that can be folded into a packet (= No Duplicates + Format: Packets) and subsequently regular labels to be placed into duplicate packets (= Duplicates Only + Format: Labels). Most herbaria donate their duplicates to other herbaria, but the herbaria receiving these specimen typically prefer using their own packets and labels. At ASU before they get distributed we store duplicates in pale blue packets, placing the label inside. Thus, when other institutions receive a donation of our specimens they can decide to print their own labels or glue the label included to their own packets, discarding the pale blue ASU duplicate packet.
______________________________________________________________________________________
Getting the Data: download CSV from the Consortium …
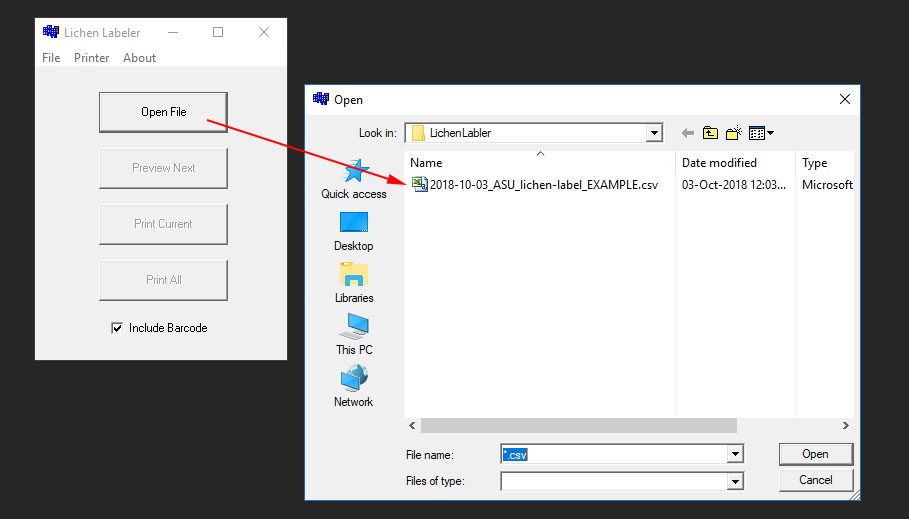
Before you start printing you will need to download your label data into a CSV and then load that file using the LichenLabler:
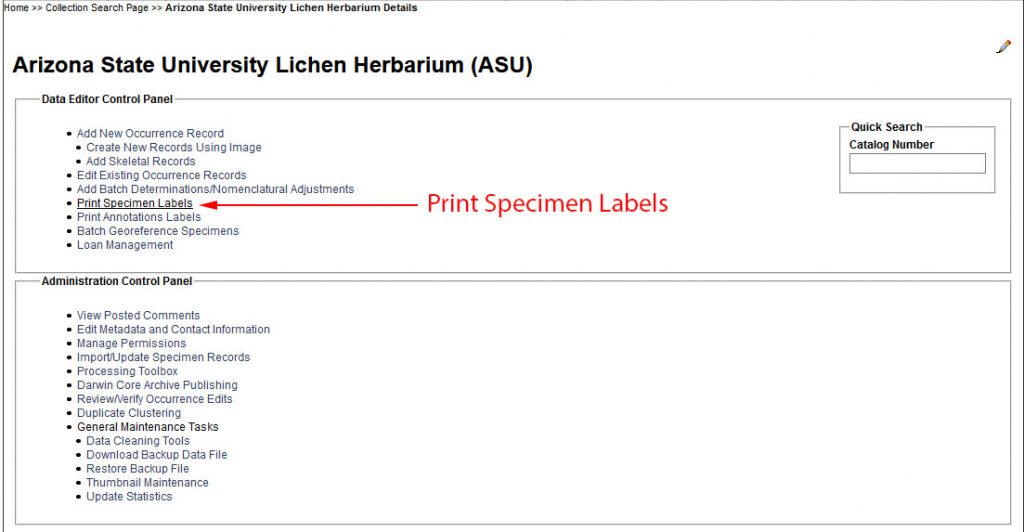
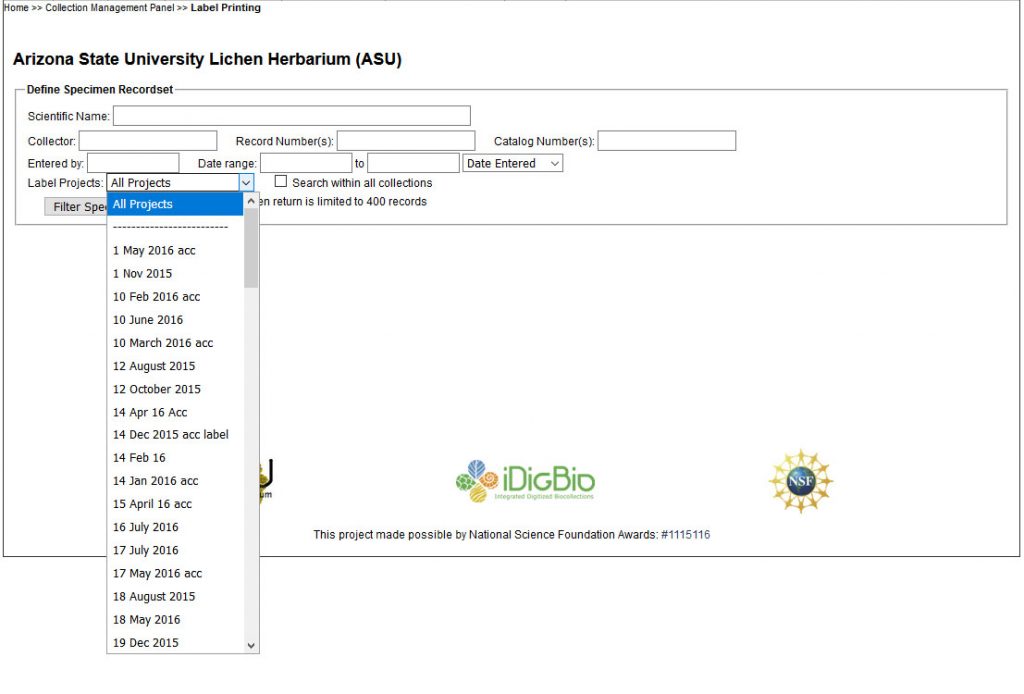
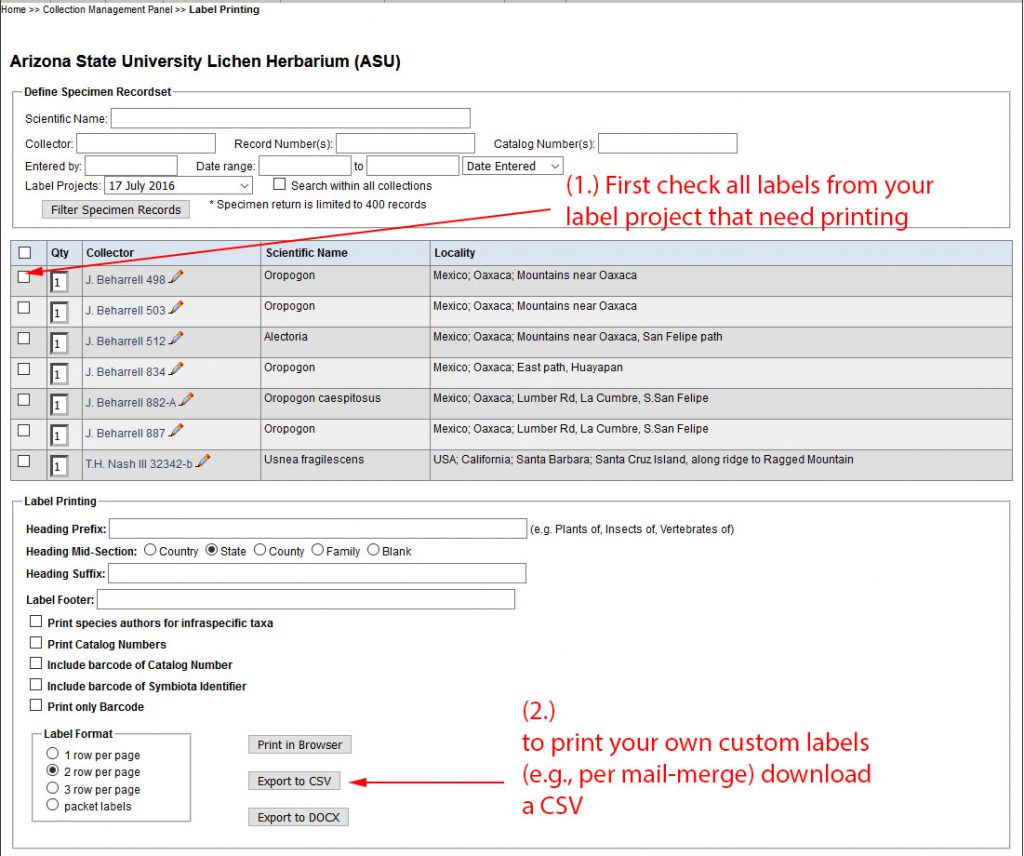
An example of what a CSV that you download looks like is included in the ZipArchive, named ASU_lichen-label_EXAMPLE.csv.
Important for Duplicate Printing:
When downloading CSV file the column labeled ‘Qty’ in the example illustrated here, includes the number of duplicate specimens. This number is the same as entered in the Duplicate field in the Occurrence Editor. However, you can manually change than number before downloading the CSV.
For each duplicate the CSV file will contain one additional row. Since version 1.9.0.0 the LichenLabler will now recognize that only the first row refers to the original specimen and subsequent rows are duplicate entries (see new configfuration options in version 1.9.0.0).
______________________________________________________________________________________
Using LichenLabler to Print Labels (all versions)
To execute the program double-click LichenLabler.exe. If you use the program frequently, place a shortcut on your desktop. On the first run you will be asked to select the printer that you want to use before a CSV file can be loaded:


______________________________________________________________________________________
This program is still in developement.
Please send any bug reports and suggestions to LichenConsortium@gmail.com
